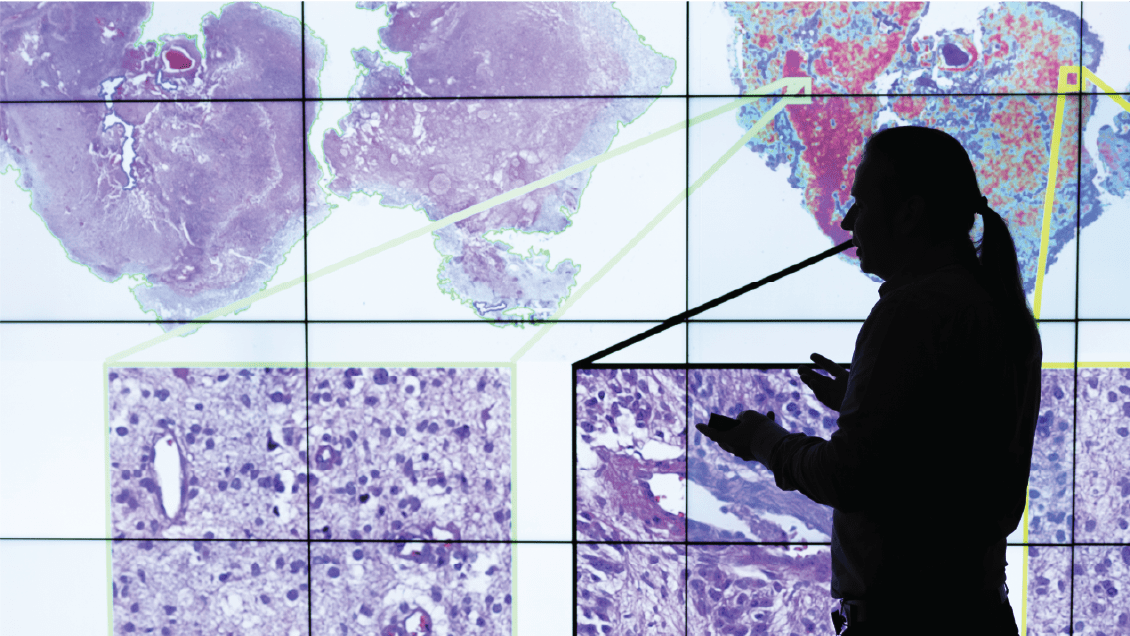
The Division of Computational Pathology, directed by Spyridon Bakas, PhD, addresses clinical requirements by developing, validating and operationalizing cutting-edge computational solutions that drive innovation in diagnostics, patient management, treatment and health care delivery, while promoting excellence in research, education and clinical care.
The division works across IU School of Medicine's tripartite mission areas: