Emerging viral diseases pose a significant threat to the health and safety of the global community. Since the beginning of the 21st century, a number of novel viruses (e.g., SARS-CoV-2) have emerged and many known viruses have become established in new locations (e.g., Zika virus). These pathogens have proven to be formidable threats to public health, leading to large outbreaks with high morbidity and, for some, high mortality. The spread of these agents has exposed many weaknesses in global infectious disease readiness and response infrastructures, and demands better diagnostics and improved countermeasures to combat them. In addition, it is essential to understand where these viruses come from and what factors govern their spillover into the human population.
A common factor in almost all outbreaks of emerging viral diseases is the involvement of one or more animal species that serve as reservoirs, intermediate hosts, and/or vectors. By investigating the relationships between zoonotic viruses and their reservoirs and hosts, it will be possible to better predict emergence so that preventive and abatement strategies can be developed and implemented in a timely fashion.
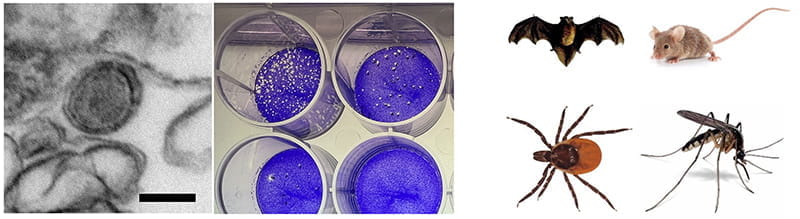
The Relich Laboratory at the Indiana University School of Medicine aims to develop accurate, precise, inexpensive, and rapidly deployable diagnostics for a number of emerging viruses. We are also interested in the ecology, epidemiology, pathogenesis, and evolution of bat-, rodent-, and arthropod-borne viruses, as well as the evaluation of novel antiviral compounds. Examples of viruses studied include Bourbon virus, chikungunya virus, dengue virus, Heartland virus, influenza viruses, Middle East respiratory syndrome coronavirus, orthohantaviruses, SARS-CoV-2 and SFTS virus.
Research is currently focused on four specific areas:
-
Development and refinement of diagnostics for emerging viruses.
The Relich Laboratory is committed to developing new technologies and methods for the diagnosis of many high-consequence viruses, including several alphaviruses, bunyaviruses, coronaviruses, flaviviruses, henipaviruses, monkeypox virus, orthomyxoviruses, and paramyxoviruses. The overarching goal of this work is to create highly sensitive, specific, and rapid diagnostic tests that are amenable to use in both laboratory and field settings. -
Surveillance for emerging pathogenic viruses in arthropods collected in Indiana and abroad.
By using a combination of classical and modern methods, we seek to determine the prevalence of several emerging viruses in possible vector and reservoir species endemic to Indiana and abroad. Work is performed at both biosafety level (BSL)-2 and BSL-3 for detection of viruses such as Bourbon virus (BRBV), California encephalitis serocomplex viruses, Heartland virus (HRTV), Lone Star virus, Powassan virus, St. Louis encephalitis virus, West Nile virus, and Western equine encephalitis virus. We are also interested in the detection and characterization of novel vector-borne viruses. -
Surveillance for emerging pathogenic viruses in bats and rodents collected in Indiana and Central America.
Similar to our arbovirus surveillance work, we are interested in detecting pathogenic viruses in wild bat and rodent populations throughout Indiana and abroad. We have partnered with the laboratory of Daniel Becker, PhD, at the University of Oklahoma to test Neotropical and domestic bats for various pathogens, especially coronaviruses. We are currently developing real-time PCR and nanopore sequencing methods to detect these viruses, and samples that yield detectable nucleic acids are inoculated onto a battery of primary and established cell lines to procure isolates for subsequent ultrastructural, biochemical, and genomic characterization. All work with potentially infectious materials is done in BSL-3 containment using enhanced safety measures to minimize potential safety risks. -
Investigating the evolution, ecology, and pathogenicity of tick-borne viruses endemic to the U.S. Midwest.
BRBV and HRTV are two relatively recently discovered tick-borne viruses that are known to be virulent human pathogens. Both have caused significant morbidity and mortality in a small number of human hosts, suggesting that these viruses, although infrequently encountered, could pose serious dangers to at-risk populations if they become more prevalent. To that end, we wish to characterize the pathogenesis of these viruses in both cell culture and in animal models to understand how they cause disease and, if possible, identify therapeutic and/or preventive countermeasures. In the future, we plan to assess BRBV and HRTV pathogenicity in collaboration with the laboratory of Natasha Tilston-Lunel, PhD at the IU School of Medicine.