DEGAS Study adds deep learning transfer to Seurat approach, creates identification of disease risk
INDIANAPOLIS–Indiana University School of Medicine and Regenstrief Institute researchers have discovered an innovative approach to identifying how likely someone is to develop certain diseases. The study, called Diagnostic Evidence GAuge of Single Cells (DEGAS), was recently published in Genome Medicine.
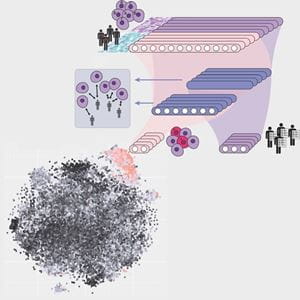
Led by Travis Johnson, PhD, assistant research professor of biostatistics and health data sciences, along with Kun Huang, PhD, MS, chair of the Department of Biostatistics and Health Data Sciences and research scientist with Regenstrief Institute, and Jie Zhang, PhD, assistant professor of medical and molecular genetics and affiliated scientist with Regenstrief Institute, the study utilized single-cell RNA sequencing that allows for the identification of differences between individual cells. The most widely used tool is known as Seurat, named after the famous French pointillism artist Georges Seurat. The sequencing tool can conjure up an image in one’s mind similar to Seurat’s style of painting, which creates images using specific points on a canvas.
Researchers took the Seurat approach, incorporated deep learning into it and named it DEGAS. Like Seraut, Edgar Degas was a famous painter. But instead of using points to create an image, Degas used impressionism.

“Incorporating a novel, deep-learning framework allowed us to transfer disease information from patients into cells,” said Johnson, the study’s lead author. “We call the transferrable information ‘impressions,’ which allowed us to assign disease attributes to individual cells.”
The team applied the DEGAS approach to Alzheimer’s disease, glioblastoma and multiple myeloma. In each disease, DEGAS allowed for deeper patient insights.
“Using the DEGAS approach allowed us to count within an individual’s single cell how many genes were present that were associated with cancer or Alzheimer’s disease and make a prediction about their risk for disease, prognosis and response to therapy,” Johnson said. “What some people don’t realize is within a single cell there are multiple genes expressed. From those genes, we may discover some associated with a variety of different cancers, neurodegenerative diseases, as well as genes associated with improved health.”
As a next step, the team plans to utilize DEGAS with spatial transcriptomics, a method that will allow them to assign cell types to their locations and identify which tumor cells and regions of cells are more associated with relapse.
This study was funded in part by the Indiana University Grand Challenge Precision Health Initiative. Additional funding was also provided by the NIH NLM-NRSA Fellowship (F31LM013056) at The Ohio State University (Columbus, OH), Multiple Myeloma Research Foundation Research Fellowship Award at Indiana University School of Medicine, and the NIH Indiana Alzheimer’s Disease Drug Discovery Center grant (U54AG065181).
Contact: Christine Drury, cldrury@iu.edu 317-385-9227 (cell)
###
About IU School of Medicine
IU School of Medicine is the largest medical school in the U.S. and is annually ranked among the top medical schools in the nation by U.S. News and World Report. The school offers high-quality medical education, access to leading medical research and rich campus life in nine Indiana cities, including rural and urban locations consistently recognized for livability.