John Wells (GS4) details his recent research in the lab of Stephanie Ware, MD, PhD, in the Department of Medical and Molecular Genetics. John is an MD/PhD student in the Medical Scientist Training Program (MSTP).
The Science
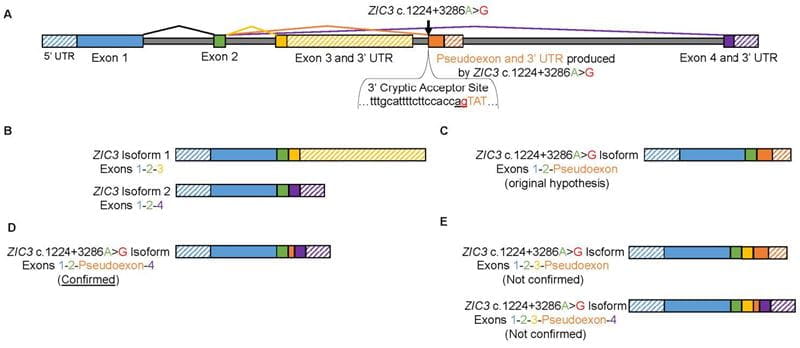
During early embryonic development, the left-right (LR) axis of our internal organs is specified by a structure known as the left-right organizer. Heterotaxy is a syndrome where individuals present with congenital heart defects and randomized LR patterning. Several genes have been reported to cause heterotaxy but mutations in the transcription factor Zinc finger of the cerebellum 3 (ZIC3) are the only known cause of X-linked heterotaxy. The ZIC3 gene is composed of 4 exons (Figure 1A) and has two known isoforms which vary based on their terminal exons. Specifically, ZIC3 Isoform 1 encodes exons 1, 2, and 3 while ZIC3 Isoform 2 encodes exons 1, 2, and 4 (Figure 1B).
Our lab identified an X-linked heterotaxy pedigree with 4 affected males. Previous lab members performed X-exome sequencing in this family and did not identify a coding mutation in ZIC3. I performed whole genome sequencing in this family and identified an intronic variant in ZIC3. This variant, ZIC3 c.1224+3286A>G, was highly predicted to alter RNA splicing by generating a cryptic 3’ splice acceptor (Figure 1A). I hypothesized this would lead to a pseudoexon inclusion (a normally intronic sequence would be mistaken as an exon) resulting in a disease associated ZIC3 c.1224+3286A>G isoform encoding exons 1, 2, and a terminal pseudoexon (Figure 1C). Patient tissue was not available to directly assess for abnormal RNA splicing. Therefore, we used CRISPR/Cas9 technology to introduce the ZIC3 c.1224+3286A>G variant into human embryonic stem cells.
My preliminary results suggest the variant does in fact alter RNA splicing. Intriguingly, the variant appears to be causing multiple abnormal splicing events simultaneously potentially resulting in multiple ZIC3 c.1224+3286A>G disease associated isoforms. Currently, one of these mutant transcripts has been confirmed by Sanger Sequencing. This disease-associated isoform encodes ZIC3 exon 1, exon 2, a pseudoexon of 57 nucleotides, and exon 4 (Figure 1D). This represents the first reported instance of a pseudoexon inclusion associated with heterotaxy for any gene. In addition, my preliminary results suggest my originally hypothesized mutant transcript (exon 1, exon 2, and a terminal pseudoexon) may still occur (Figure 1C), but this has not yet been confirmed. Furthermore, I now suspect additional mutant transcripts may include a portion of ZIC3 exon 3 (Figure 1E).
Although I have now shown the ZIC3 c.1224+3286A>G variant does in fact lead to a pseudoexon inclusion, I now have several new questions: How many mutant isoforms are there? What do they encode? What are their expression levels relative to the normal ZIC3 isoforms?
The Future
My current career goal is to become a clinical geneticist in an academic setting. As a physician, I hope to make an impact at an individual level. I want to help identify the causes of genetic syndromes in my patients and potentially treat them using methods such as gene therapy or antisense oligonucleotide therapy. As a scientist, I hope my research accomplishments can benefit the field of genetics as a whole. For example, my exemplary finding discussed here suggests genetic researchers should consider non-coding variants and novel mechanisms of disease (Ex: irregular RNA splicing, abnormal post-transcriptional modification, or disruption of gene-regulatory networks) in their unsolved cases.
